Analysis
New AI system designs proteins that efficiently bind to focus on molecules, with potential for advancing drug design, illness understanding and extra.
Each organic course of within the physique, from cell development to immune responses, is determined by interactions between molecules known as proteins. Like a key to a lock, one protein can bind to a different, serving to regulate important mobile processes. Protein construction prediction instruments like AlphaFold have already given us large perception into how proteins work together with one another to carry out their capabilities, however these instruments can’t create new proteins to immediately manipulate these interactions.
Scientists, nevertheless, can create novel proteins that efficiently bind to focus on molecules. These binders may also help researchers speed up progress throughout a broad spectrum of analysis, together with drug improvement, cell and tissue imaging, illness understanding and analysis – even crop resistance to pests. Whereas latest machine studying approaches to protein design have made nice strides, the method remains to be laborious and requires in depth experimental testing.
Immediately, we introduce AlphaProteo, our first AI system for designing novel, high-strength protein binders to function constructing blocks for organic and well being analysis. This expertise has the potential to speed up our understanding of organic processes, and help the invention of recent medicine, the event of biosensors and extra.
AlphaProteo can generate new protein binders for various goal proteins, together with VEGF-A, which is related to most cancers and problems from diabetes. That is the primary time an AI software has been in a position to design a profitable protein binder for VEGF-A.
AlphaProteo additionally achieves greater experimental success charges and three to 300 instances higher binding affinities than one of the best current strategies on seven goal proteins we examined.
Studying the intricate methods proteins bind to one another
Protein binders that may bind tightly to a goal protein are onerous to design. Conventional strategies are time intensive, requiring a number of rounds of intensive lab work. After the binders are created, they bear further experimental rounds to optimize binding affinity, so that they bind tightly sufficient to be helpful.
Skilled on massive quantities of protein knowledge from the Protein Knowledge Financial institution (PDB) and greater than 100 million predicted constructions from AlphaFold, AlphaProteo has realized the myriad methods molecules bind to one another. Given the construction of a goal molecule and a set of most well-liked binding areas on that molecule, AlphaProteo generates a candidate protein that binds to the goal at these areas.
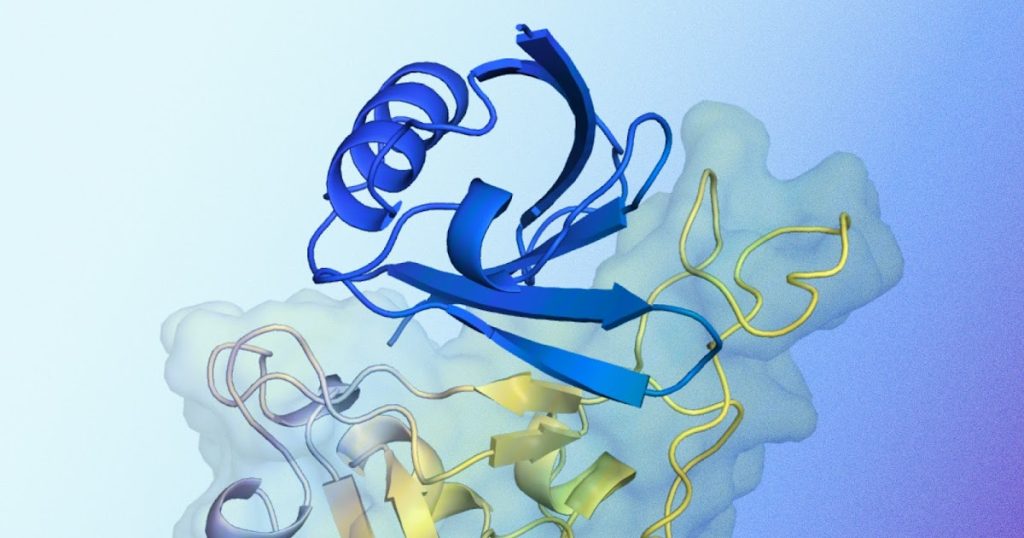
Illustration of a predicted protein binder construction interacting with a goal protein. Proven in blue is a predicted protein binder construction generated by AlphaProteo, designed for binding to a goal protein. Proven in yellow is the goal protein, particularly the SARS-CoV-2 spike receptor-binding area
Demonstrating success on vital protein binding targets
To check AlphaProteo, we designed binders for various goal proteins, together with two viral proteins concerned in an infection, BHRF1 and SARS-CoV-2 spike protein receptor-binding area, SC2RBD, and 5 proteins concerned in most cancers, irritation and autoimmune illnesses, IL-7Rɑ, PD-L1, TrkA, IL-17A and VEGF-A.
Our system has highly-competitive binding success charges and best-in-class binding strengths. For seven targets, AlphaProteo generated candidate proteins in-silico that sure strongly to their supposed proteins when examined experimentally.
A grid of illustrations of predicted constructions of seven goal proteins for which AlphaProteo generated profitable binders. Proven in blue are examples of binders examined within the moist lab, proven in yellow are protein targets, and highlighted in darkish yellow are supposed binding areas.
For one explicit goal, the viral protein BHRF1, 88% of our candidate molecules sure efficiently when examined within the Google DeepMind Moist Lab. Primarily based on the targets examined, AlphaProteo binders additionally bind 10 instances extra strongly, on common, than one of the best current design strategies.
For an additional goal, TrkA, our binders are even stronger than one of the best prior designed binders to this goal which have been by means of a number of rounds of experimental optimization.
Bar graph displaying experimental in vitro success charges of AlphaProteo’s output for every of the seven goal proteins, in comparison with different design strategies. Larger success charges imply fewer designs have to be examined to seek out profitable binders.
Bar graph displaying one of the best affinity for AlphaProteo’s designs with out experimental optimization for every of the seven goal proteins, in comparison with different design strategies. Decrease affinity means the binder protein binds extra tightly to the goal protein. Please be aware the logarithmic scale of the vertical axis.
Validating our outcomes
Past in silico validation and testing AlphaProteo in our moist lab, we engaged Peter Cherepanov’s, Katie Bentley’s and David LV Bauer’s analysis teams from the Francis Crick Institute to validate our protein binders. Throughout totally different experiments, they dived deeper into a few of our stronger SC2RBD and VEGF-A binders. The analysis teams confirmed that the binding interactions of those binders had been certainly much like what AlphaProteo had predicted. Moreover, the teams confirmed that the binders have helpful organic operate. For instance, a few of our SC2RBD binders had been proven to stop SARS-CoV-2 and a few of its variants from infecting cells.
AlphaProteo’s efficiency signifies that it might drastically cut back the time wanted for preliminary experiments involving protein binders for a broad vary of purposes. Nonetheless, we all know that our AI system has limitations, because it was unable to design profitable binders in opposition to an eighth goal, TNFɑ, a protein related to autoimmune illnesses like rheumatoid arthritis. We chosen TNFɑ to robustly problem AlphaProteo, as computational evaluation confirmed that it might be extraordinarily tough to design binders in opposition to. We are going to proceed to enhance and develop AlphaProteo’s capabilities with the purpose of finally addressing such difficult targets.
Attaining robust binding is normally solely step one in designing proteins that may be helpful for sensible purposes, and there are a lot of extra bioengineering obstacles to beat within the analysis and improvement course of.
In direction of accountable improvement of protein design
Protein design is a fast-evolving expertise that holds a lot of potential for advancing science in all the pieces from understanding the components that trigger illness, to accelerating diagnostic check improvement for virus outbreaks, supporting extra sustainable manufacturing processes, and even cleansing contaminants from the atmosphere.
To account for potential dangers in biosecurity, constructing on our long-standing strategy to accountability and security, we’re working with main exterior consultants to tell our phased strategy to sharing this work, and feeding into group efforts to develop finest practices, together with the NTI’s (Nuclear Risk Initiative) new AI Bio Discussion board.
Going ahead, we’ll be working with the scientific group to leverage AlphaProteo on impactful biology issues and perceive its limitations. We have additionally been exploring its drug design purposes at Isomorphic Labs, and are excited for what the longer term holds.
On the identical time, we’re persevering with to enhance the success price and affinity of AlphaProteo’s algorithms, increasing the vary of design issues it may sort out, and dealing with researchers in machine studying, structural biology, biochemistry and different disciplines to develop a accountable and extra complete protein design providing for the group.
In the event you’re a biologist, whose analysis may gain advantage from target-specific protein binding, and also you’d wish to register curiosity in being a trusted tester for AlphaProteo, please attain out to us on alphaproteo@google.com.
We’ll course of messages obtained in accordance with our Privateness Coverage.
Acknowledgements
This analysis was co-developed by our Protein Design crew and Moist Lab crew.
We’d wish to thank our collaborators Peter Cherepanov, David Bauer, Katie Bentley and their teams on the Francis Crick Institute for his or her invaluable experimental insights and outcomes, the AlphaFold crew, whose earlier work and algorithms offered coaching inputs and analysis insights, and the various different groups throughout Google DeepMind who contributed to this program.